P3DIA – Unlocking the Non-Coding Genome
Prioritization of Genome Data with Image Analysis for non-coding and regulatory variants
Why this study?
Even with whole genome sequencing (GS), in 50–60% of patients the causal variant remains undetected. A major reason is that non-coding and regulatory variants are difficult to interpret, and standard analysis pipelines often fail to bring them to the top of the candidate list.
What is P3DIA?
P3DIA (Prioritization of Genome Data with Image Analysis) is a large, prospective, multicenter study within the Modellvorhaben Genomsequenzierung (MV-GenomeSeq) framework.
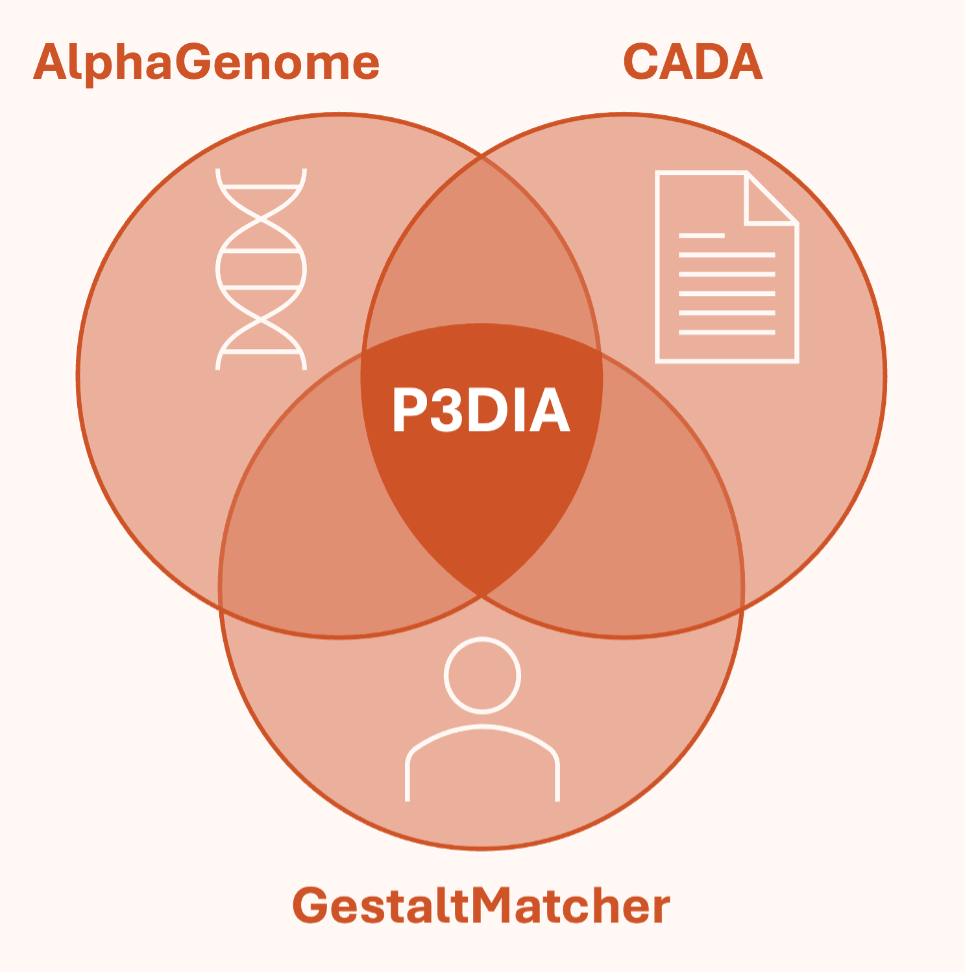
An integrated AI pipeline combining three complementary evidence sources:
- Sequence-based (AlphaGenome: regulatory + splicing impact)
- Image-based (GestaltMatcher: facial features)
- Phenotype-based (CADA-prio: HPO-driven gene fit)
This multimodal approach aims to improve variant prioritization, especially when the causal variant lies outside coding regions.
The PEDIA studies – From proof-of-concept to P3DIA
P3DIA is the third generation of the PEDIA project and is pronounced simply as “Pedia Three”. The “3” marks this third phase, building directly on the foundations of PEDIA 1 and PEDIA 2. Each step expanded in scope and ambition: from early proof-of-concept, to clinical validation, and now to large-scale implementation of multimodal phenotyping for non-coding variants.
| Feature | PEDIA 1 Hsieh et al., Genet Med 2019 |
PEDIA 2 Schmidt et al., Nat Genet 2024 |
P3DIA |
|---|---|---|---|
| Duration | 2017–2019 | 2019–2024 | 2025–2029 |
| Primary utility | Proof-of-concept | Validation in clinical practice | Scaling & expansion |
| Study type | Retrospective (simulation) | Prospective, multicenter | Prospective, multicenter |
| Sequencing | Exome (simulated) | Clinical exome | Genome (Trio) |
| Sites | – | 10 clinics | >25 university clinics & diagnostic labs |
| Image data | 2D | 2D | 2D + 3D |
| Family inclusion | – | Patients only | Trio + ≥1 unaffected person |
Together, the PEDIA studies show the evolution from concept → validation → scaling, with P3DIA now taking the next step to unlock the non-coding genome.
Frequently Asked Questions
Who can participate?
We welcome collaborators who can contribute rich datasets and phenotypic information:
- Images of the trio (mother, father, child)
- Longitudinal images (patient at different ages)
- Additional modalities (X-ray, MRI, fundoscopies…)
- 3D scan of the patient with the new GestaltMatcher 3D app
On which platforms will P3DIA prioritization be available?
We will initially implement P3DIA within Emedgene, since the majority of centers in MV-GenomeSeq use Illumina sequencers. However, the pipeline is platform-agnostic and can be integrated with minor adjustments into tertiary analysis tools such as VarSome or enGenome eVai.
What’s in it for collaborators?
- Prioritization of non-coding variants in unresolved cases
- Early access to phenotype-driven analysis
- Co-authorship opportunities on resulting publications
How do we support you?
We can provide:
- Assistance with ethics applications
- Technical support for data integration
- Flexible participation models
Get Involved
For more information or collaboration opportunities, please contact us by email using the subject line “P3DIA”.
Prof. Peter Krawitz
Institute for Genomic Statistics and Bioinformatics
University Clinic Bonn
✉️ pkrawitz@uni-bonn.de
Dr. Adele Ruder
Institute for Genomic Statistics and Bioinformatics
University Clinic Bonn
✉️ ruder@uni-bonn.de